The new look
Biodiversity Heritage Library has just launched. It's a complete refresh of the old site, based on the
Biodiversity Heritage Library–Australia site. If you want an overview of what's new, BHL have published a guide to the
new look site. Congrats to involved in the relaunch.
One of the new features draws on the work I've been doing on
BioStor. The new BHL interface adds the notion of "parts" of an item, which you can see under the "Table of Contents" tab. For example, the scanned
volume 109 of the Proceedings of the Entomological Society of Washington now displays a list of articles within that volume:
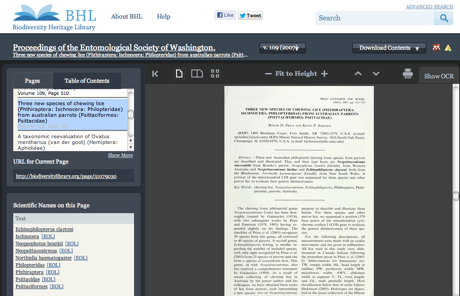
This means you can now jump to individual articles. Before you had to scroll through the scan, or click through page numbers until you found what you were after. The screenshot above shows the article "Three new species of chewing lice (Phthiraptera: Ischnocera: Philopteridae) from australian parrots (Psittaciformes: Psittacidae)". The details of this article have been extracted from BioStor, where this article appears as
http://biostor.org/reference/55323. You can go directly to this article in BHL using the link
http://www.biodiversitylibrary.org/part/69723. As an aside, I've chosen this article because it helps demonstrate that BHL has modern content as well as pre-1923 literature, and this article names a louse,
Neopsittaconirmus vincesmithi after a former student of mine,
Vince Smith. You're nobody in this field unless you've had a louse named after you ;)
BioStor has over 90,000 articles, but this is a tiny fraction of the articles contained in BHL content, so there's a long way to go until the entire archive is indexed to article level. There will also be errors in the article metadata derived from BioStor. If we invoke Linus's Law (
"given enough eyeballs, all bugs are shallow") then having this content in BHL should help expose those errors more rapidly.
As always, I have a few niggles about the site, but I'll save those for another time. For noe, I'm happy to celebrate an extraordinary, open access archive of over 40 million pages. BHL represents one of the few truly indispensable biodiversity resources online.

